Statistical analysis of biological sequences
Biological sequences and how to represent them.
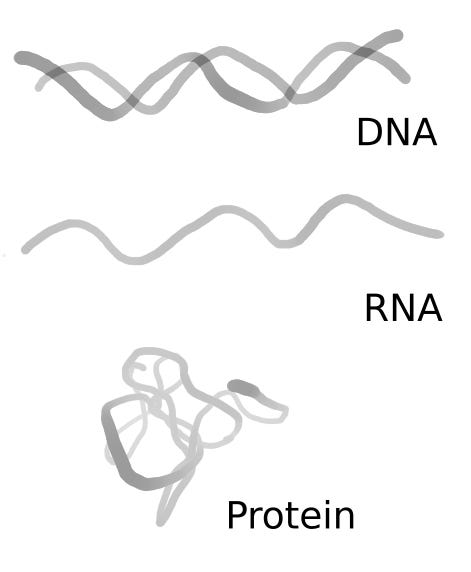
Biological sequences are perhaps the main source of information in biological systems. In simple terms, a biological sequence is a series of ordered elements, and the kind of elements determine the kind of sequence. There are at least three kinds of biological sequences, DNA, RNA, and protein sequences. Those three kinds of sequences are closely related as DNA is transcribed to RNA and then translated to protein. Roughly DNA/RNA are the blueprints to design the different components that make a cell. While proteins are the components that organize and perform different tasks that make a cell work.
Component wise DNA/RNA share more similarities compared to proteins. DNA is made by deoxynucleotides, RNA by ribonucleotides, and proteins by amino acids. DNA/RNA sequences are longer compared to proteins, and the number of unique elements is only four. Contrary to DNA/RNA, proteins are generally shorter in size, and the number of possible unique elements is 21. One of the main reasons for this difference is that DNA and RNA encode the information of the different amino acids as a combination of nucleotides. Using the combination reduces the number of unique elements but increases its size.
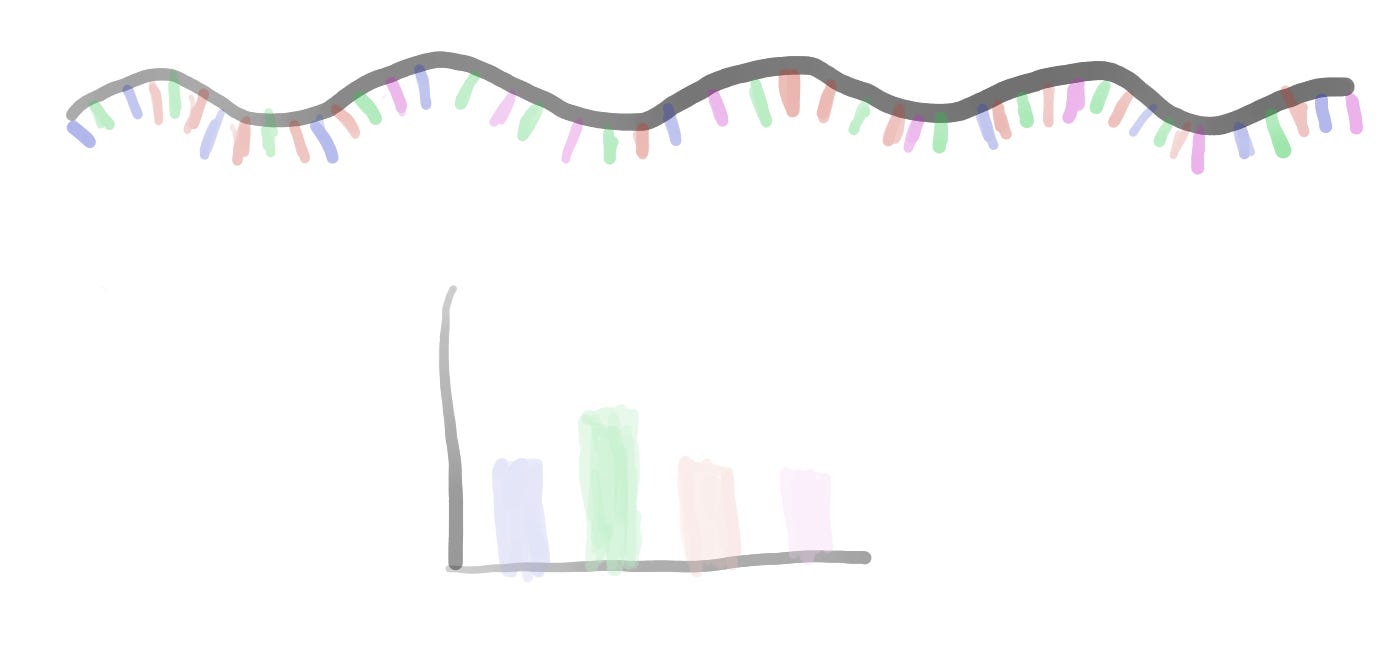
The first and perhaps the most common representation of a sequence is a continuous string. Where each letter represents an element and the order in the string is the same as the one in the biological sequence. This representation preserves the order of the sequence yet its size could make it difficult to analyze. A short-size alternative is the use of element frequency. It also has the advantage of having a chemical meaning as it represents the number of elements needed to make the sequence.
Viral sequences and time series.
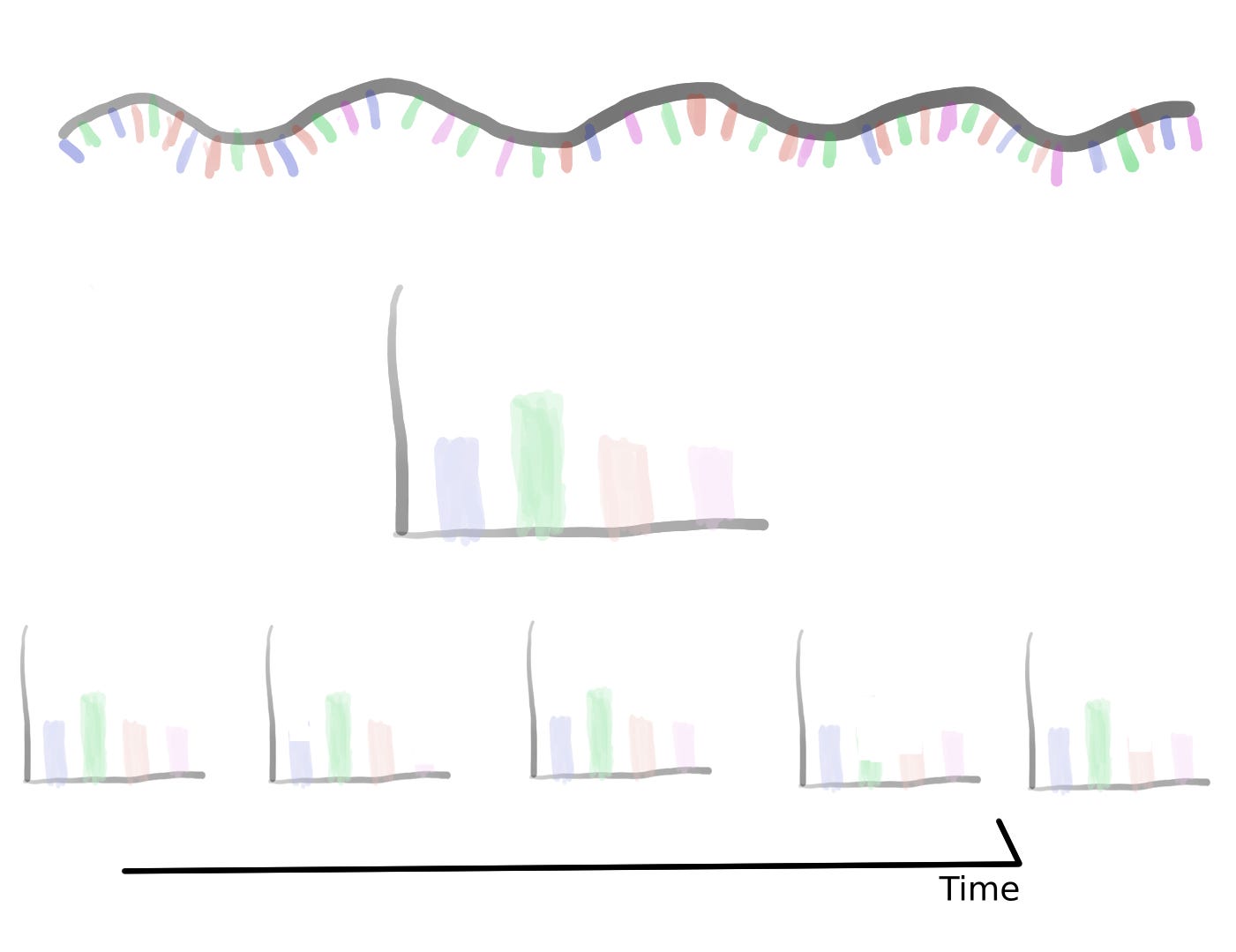
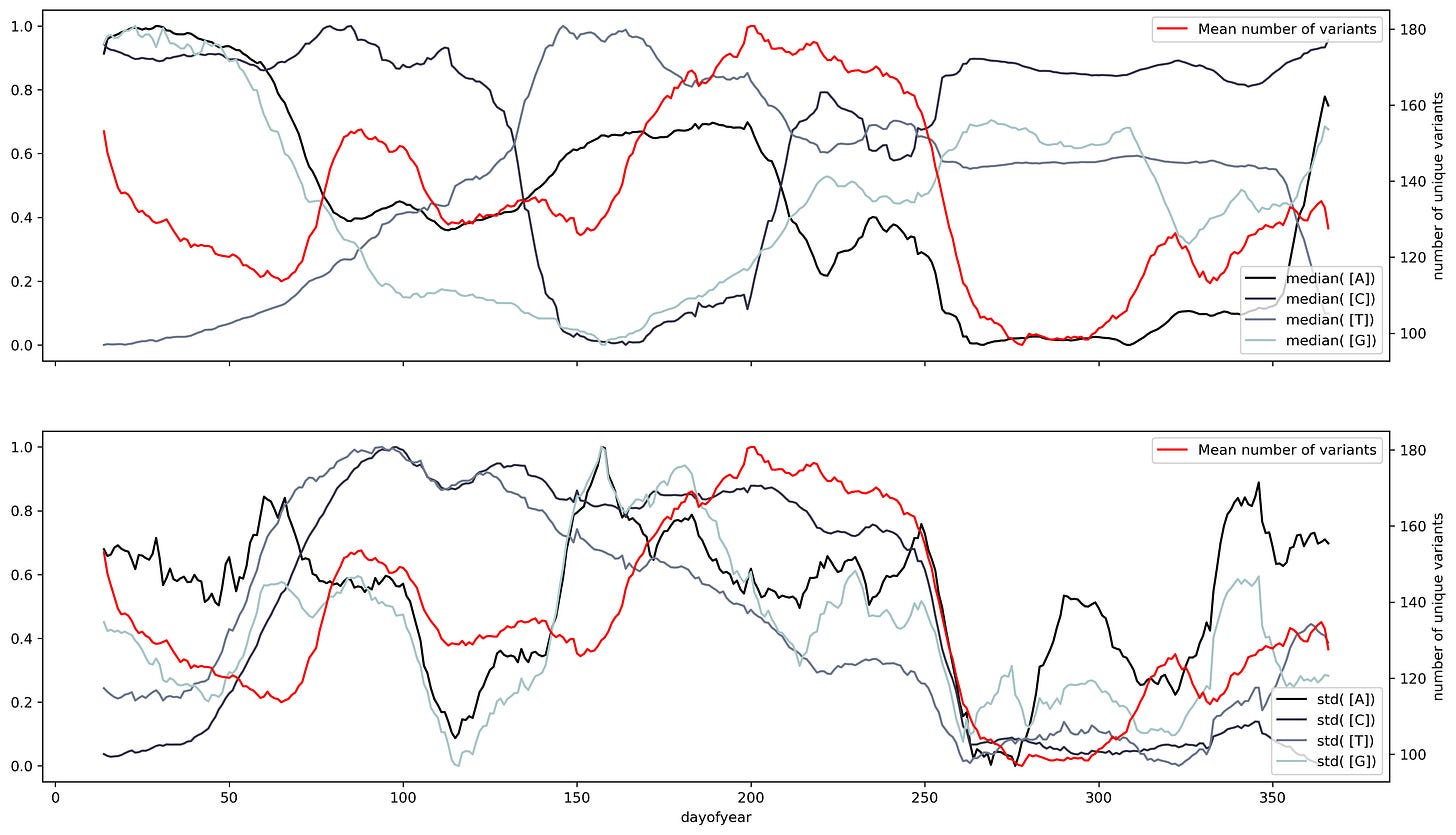
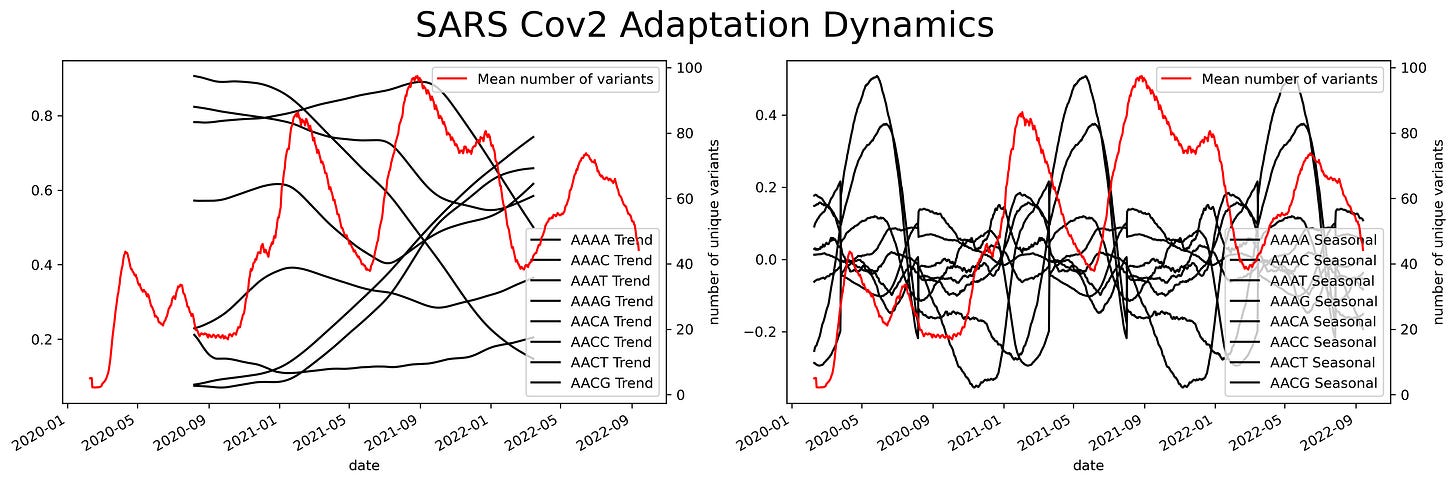
Having a small-size representation of the sequence allows us to use large databases of biological sequences. Currently, due to genomic surveillance, there’s a great number of SARS-Cov2 sequences openly available and ready to be analyzed. We can use the frequency-based representation and the isolation date of the sequence to construct a time series. This will allow us to assess small variations through time along the ribonucleotides that make the SARS-Cov2 genome. But, too much variation could hinder our ability to make a clear assessment of the data. To account for variability we can use a sliding window.
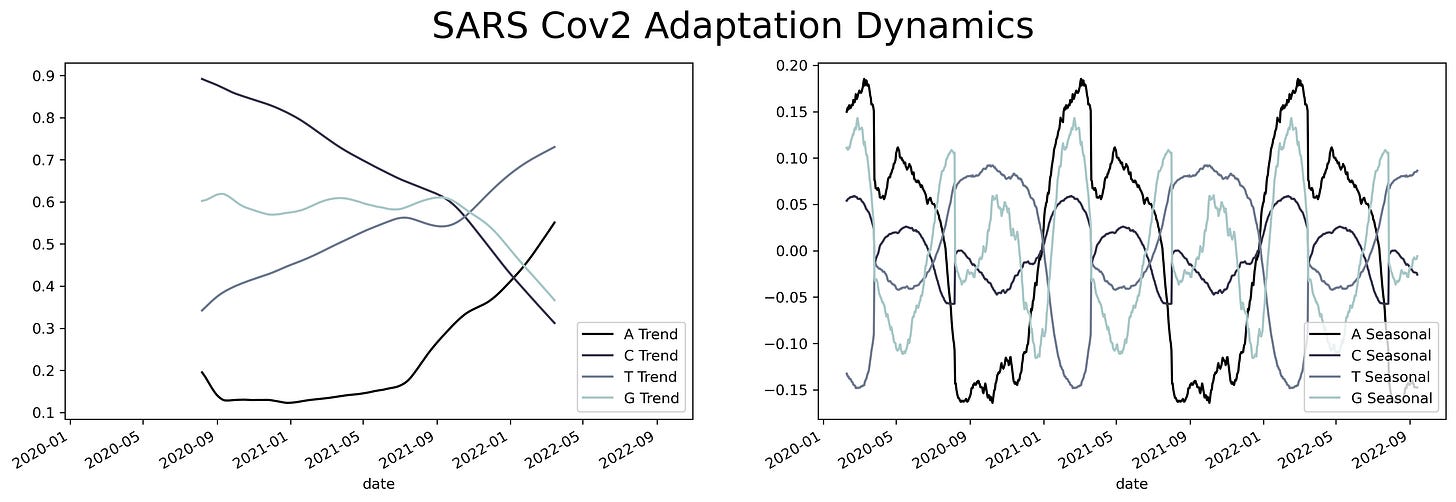
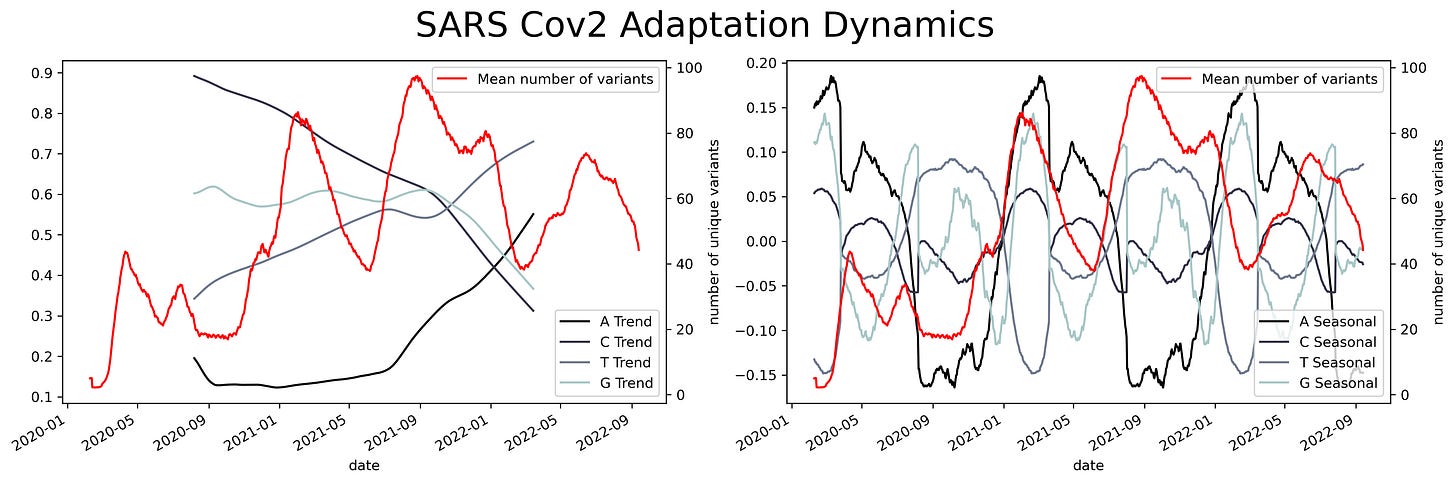
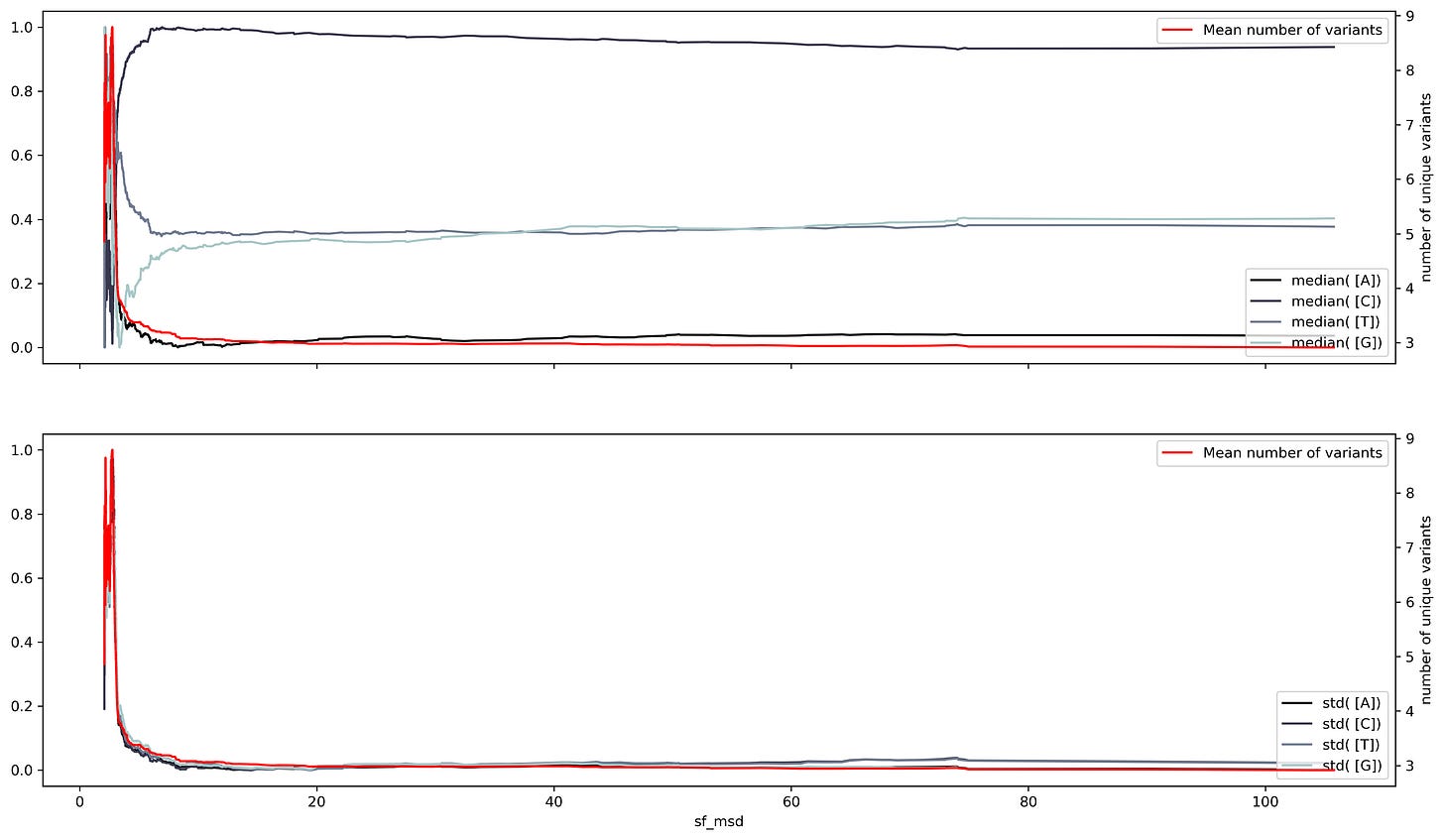
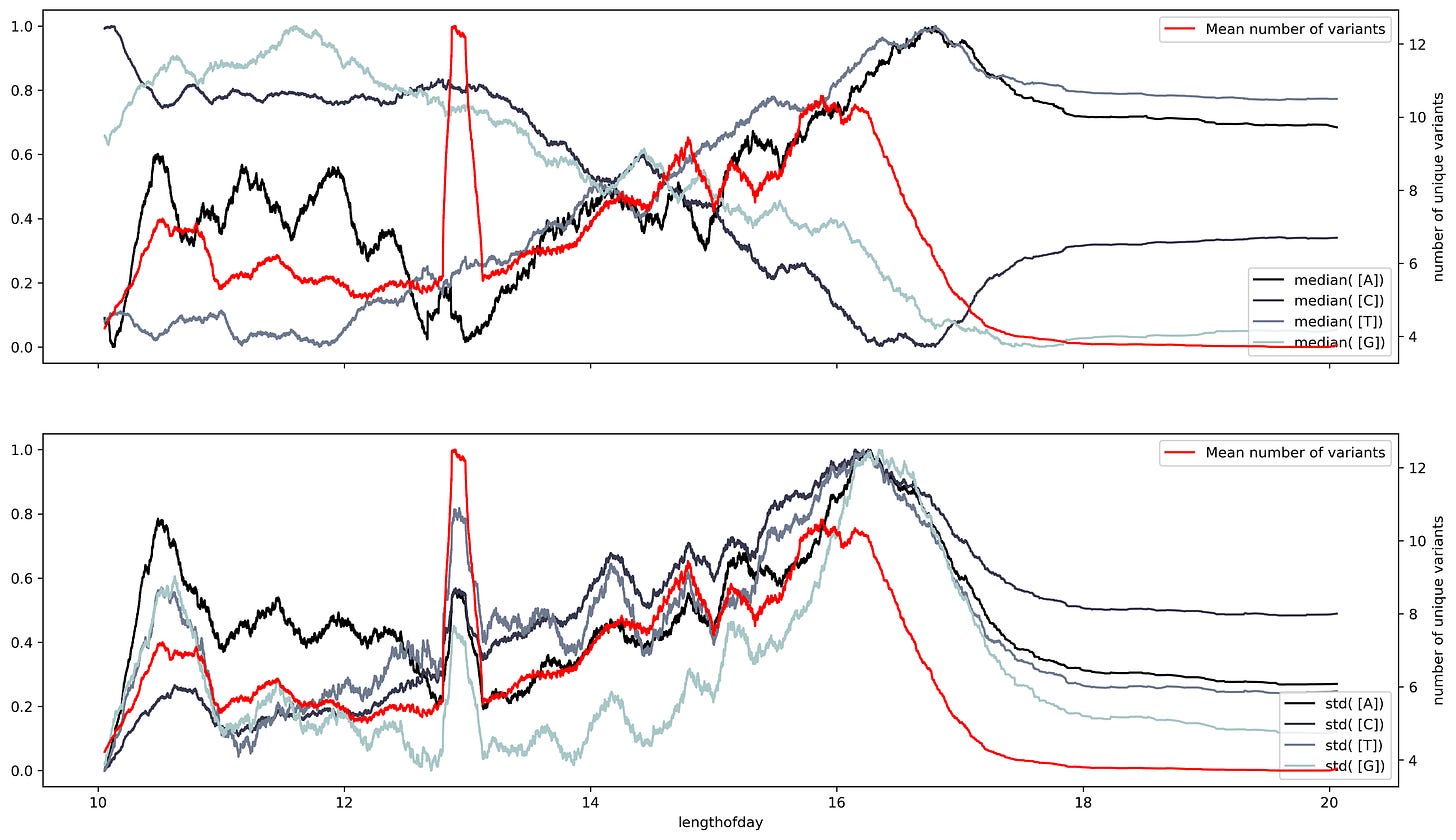
A sliding window consists of taking a fragment of the time series and estimating some statistics. Then we slide a single unit of time and calculate the same statistic to a new fragment of the same size. This procedure is applied to the entire time series resulting in a new time series with less noise than the original. Appling that analysis to the SARS Cov2 sequences results in a series of time series with several oscillations through time.
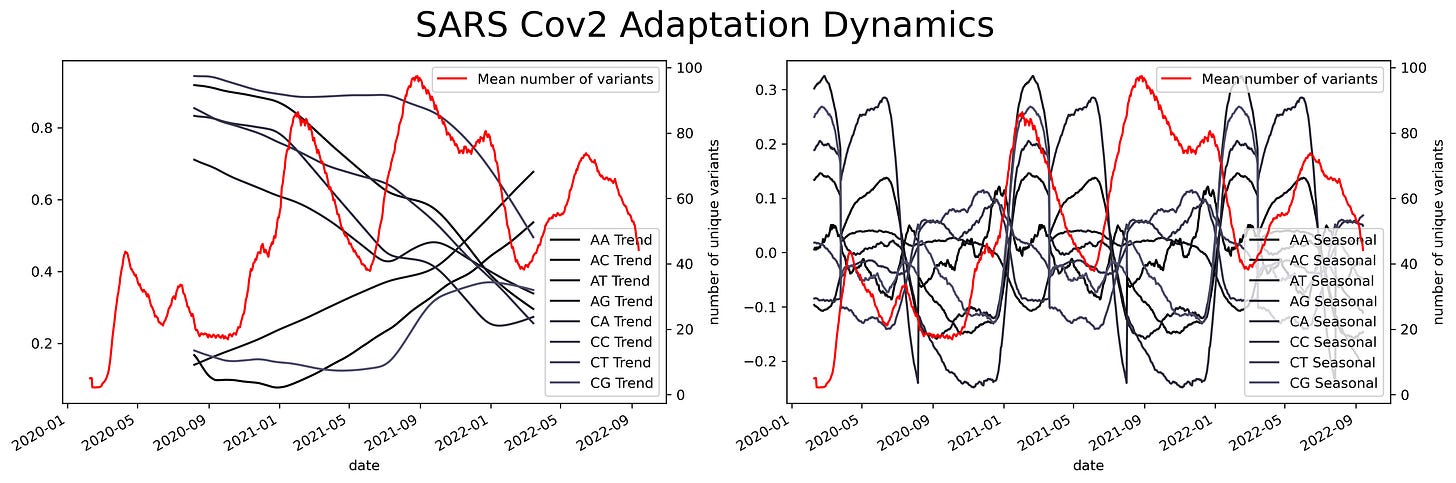
Oscillations could have a variety of meanings, they could mean mutational hotspots or infectivity hotspots. To have a better understanding of that meaning the time series can be decomposed into two components, a trend component, and a seasonal component. The trend component will contain information about the long-term change of the SARS-Cov2 sequence or an increase in a specific ribonucleotide. This characteristic can be used to design long-term treatments, as analogs to nucleotides are often used as antiviral treatment. An increase in the use of a specific nucleotide will point towards a resource dependence on that ribonucleotide. Making the virus more susceptible to that specific nucleotide.
While the seasonal component could be helpful to predict future COVID-19 waves or mutational hotspots. Assessing a possible correlation between mutations in the virus and the seasonal component can be done by plotting the isolated variants and the different components. This shows a slight overlap between the seasonal components and the variants. Particularly at times were there's a shift in the nucleotide content.
Mixed time scales
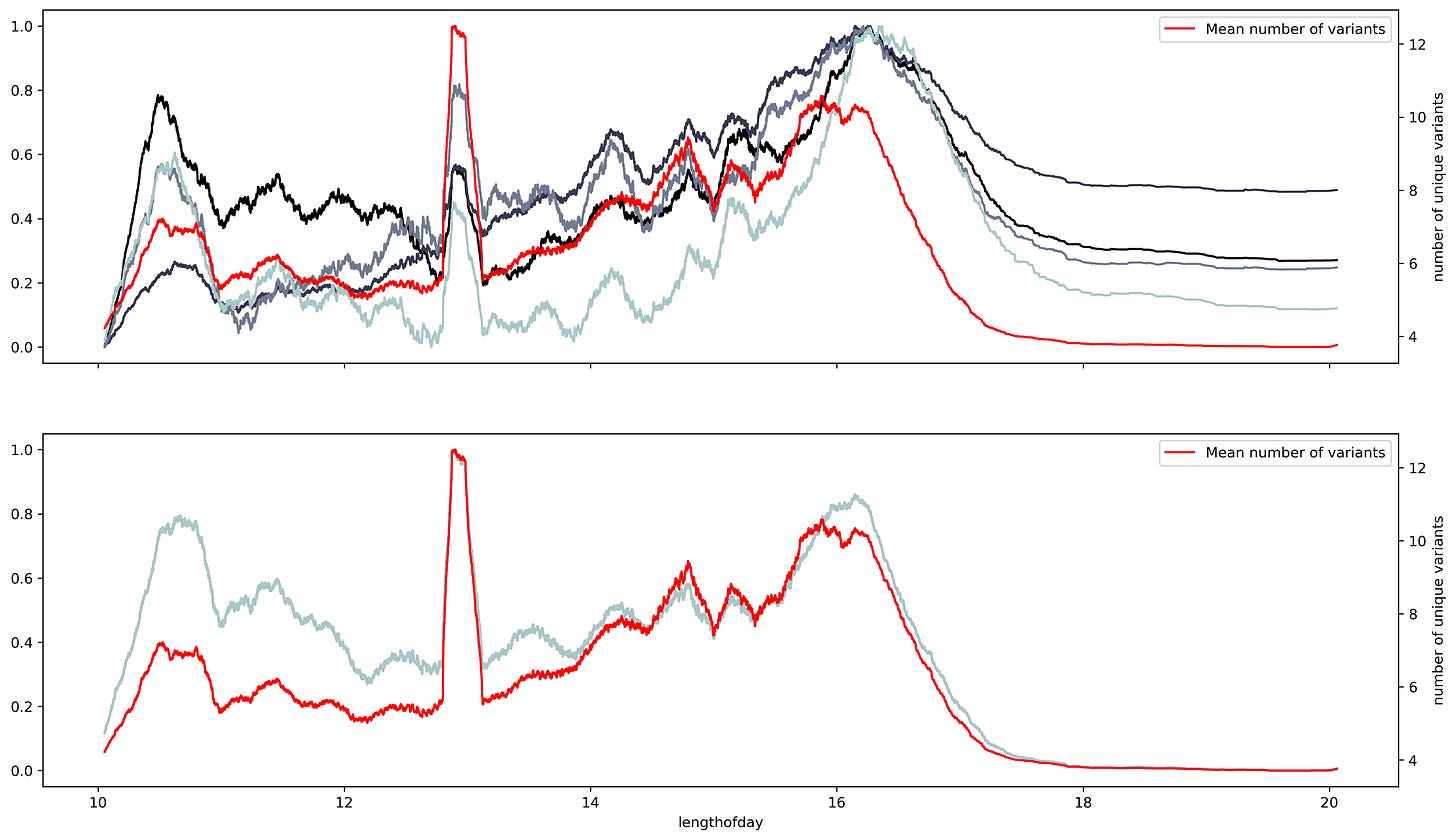
Changing the timescale used to make the time series to yearly scale results in overlapping between the variants and shift regions. However, the overlap appears to have a better match to the standard deviation. This makes sense, the standard deviation is a dispersion measure, and more variants will result in a greater dispersion.
But to evaluate the relationship between variants and the ribonucleotide change the different seasonal components need to be synchronized. There should be a way to arrange the sequences where the seasonal component starts at the same time for all the ribonucleotides. Currently, the seasonality of COVID-19 waves is unknown. But, there's a great deal of evidence pointing towards at least two factors, solar radiation, and latitude. A simple form to combine both attributes is by measuring the sunshine duration or the solar flux. The solar flux refers to the amount of radiation at a particular location. The solar flux results in a complicated pattern, while, the sunshine duration results in a profile very like the number of isolated variants.
Although the match is not perfect it points out that dispersion measures reflect better the dependence between ribonucleotide content and the number of variants. Another dispersion measurement is entropy, which measures the amount of information within a variable. Entropy measurements show an almost perfect fit between the number of variants and changes in entropy per ribonucleotide. Suggesting that the number of variants is an indirect measurement of the entropy within the SARS-Cov2 sequence at a given time. Also that the SARS-Cov2 seasonality must show a large correlation between the number of variants and the entropy of the sequence. Thus the sunshine duration offers an approximate time scale to asses the seasonality of the virus, yet, is not exact.
Sequence sliding sampling
Up to this point, we have only used the frequency of single ribonucleotides as a method for sequence representation. Although part of the information is lost, the different analysis techniques already show that is possible to get valuable information from this simple representation. But the sequence representation scheme can be expanded by borrowing the sliding sampling from the time series analysis. The sequence is then divided into fragments of size k by sliding through the sequence and then the frequency of each of the fragments is used as a small-size representation of the sequence.
Then the previous analysis is applied to the new representation. However, the new sampling scheme results in a large number of unique elements. By taking a single ribonucleotide there are only four unique elements, taking two ribonucleotides results in 16 unique combinations and the number continues t rise. Taking a sliding window of size two and applying the same analysis results in a series of elements with perhaps a better linearization on the trend component.
But taking a sliding window of size four results in 256 different combinations of unique fragments. This large number of fragments complicates the ability to get any kind of information from the analysis. This shows the necessity to develop more accurate methods to analyze the fragment-based representation of sequences. In the next posts, I will show you other methods to analyze and classify sequences using fragment-based representation.
Some extra notes
Even when it seems that the sunshine duration has no relationship with COVID-19 its use wasn't random. It provides an easy way to combine solar radiation and latitude. On one side vitamin D (a vitamin synthesized due to UV radiation) deficiency has been linked to a higher risk of complications due to COVID-19. Also, IR radiation treatment has been found to increase the recovery time of non-hospitalized COVID-19 patients. Further IR radiation is also able to inactivate the toxic effects of the spike protein. Although the exact mechanism a clear relationship exists between solar radiation and COVID-19.
Vitamina D
Autumn COVID‑19 surge dates in Europe correlated to latitudes, not to temperature‑humidity, pointing to vitamin D as contributing factor.
Radiation COVID-19
Cardiopulmonary and hematological effects of infrared LED photobiomodulation in the treatment of SARS-COV2
Radiation Spike
Infrared light therapy relieves TLR-4 dependent hyper-inflammation of the type induced by COVID-19
The Plan
The main objective of this blog is to raise funds for the further development of the methods described in Applications of sliding sampling to biological sequences, as well as a series of posts on Medium. Further development of such methods will fast-track the characterization of emerging pathogens and other diseases. Data, code, and examples of how to use the methodology will be periodically published and will remain open.
The development of scientific research outside of academia is a growing movement known as citizen science. However, funding of such projects continues to be a limiting factor. Supporting this kind of project will new ideas for a scientific problem.
Avoid large crowds at times near stationary points of solar radiation, noon, winter, and summer. Increase your daily exposure to solar radiation. And see you in the next one.